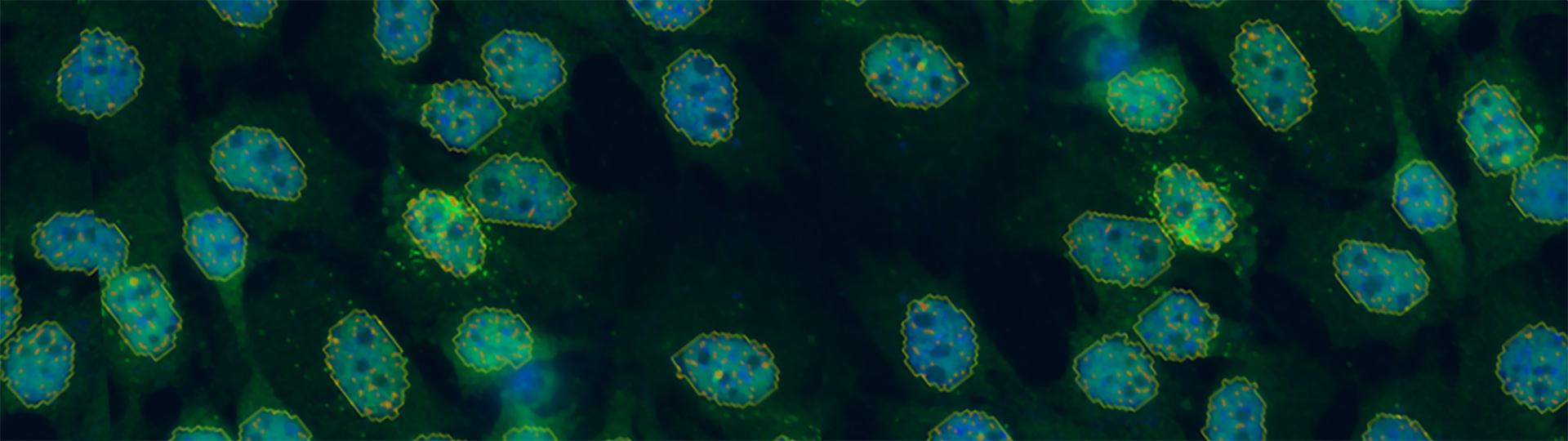

Single Molecule FISH (smFISH) Imaging
A Fluorescence Localization and Imaging Technique for RNA Visualization
RNA transcripts are a critical piece of the central dogma of biology yet are difficult to image due to their requirement for specific, fluorescent labeling and nanoscopic size. Bruker’s Vutara single-molecule localization microscope (SMLM) and fully integrated SRX software enables users to easily conduct smFISH imaging and analysis experiments.
Bruker offers the tools necessary for successful smFISH and super-resolution experiments including our:
- Vutara VXL SMLM microscope with localization capabilities;
- User-friendly SRX software module optimized for smFISH imaging and analysis; and
- Microfluidics unit for multiplexing of smFISH experiments requiring a large number of fluorescent probes.
What is Single Molecule FISH (smFISH)?
Single molecule fluorescence in situ hybridization (smFISH) — also known as smRNA FISH or RNA FISH — is a cutting-edge technique for studying gene expression in single cells. This technique is similar to FISH in that it is used to visualize DNA, either specific genes or portions of genes, but differs in its unique ability to image and quantify individual RNA molecules. The applications of smFISH transcend life sciences disciplines and include, among others, genomics, cancer biology, neuroscience, and more.
Key advantages of smFISH
- Enables visualization and quantification of specifically labeled RNA molecules at single-molecule resolution; and
- Supports single-cell gene expression experiments through time and space.
P E O P L E A L S O A S K:
If the research question requires investigation and visualization of RNA instead of DNA, smFISH is the most optimized technique. Fluorescence in-situ hybridization (FISH) operates under the same principles as smFISH — the hybridization of complementary fluorescent probes to a target sequence. However, FISH is optimized for targeting DNA sequences, such as entire genes or portions of genes.
Visit our Super-Resolution Microscopy Knowledge Pack, which contains a bundle of information. Inside, you will find the Bruker Super-Resolution eBook, several full-length protocol papers, and a full-length webinar. These resources are freely available and can be accessed on the Super-Resolution Microscopy Knowledge Pack page.
What is smFISH Used For?
Visualize and quantify specifically labeled RNA with single-molecule resolution
Performing smFISH experiments with the Vutara not only allows for quantification of RNA, but also enables the visualization of the target RNA at subcellular resolution. The ability to image the RNA within the cell opens avenues to studying RNA distribution and localization in relation to other markers or proteins. The combination of the imaging and quantification with smFISH can also be used as a readout for prediction of protein content. The advantage of RNA imaging with smFISH with the Vutara is unique from RNA sequencing data, which only provides quantitative values of RNA abundance.
Investigate spatio-temporal gene expression patterns
RNA imaging with smFISH enables quantitative and qualitative gene expression experiments within single cells. With smFISH, researchers can measure cellular mRNA and perform spatial localization and quantification in locations such as the cytoplasm and nucleus. Single RNA molecules can be traced over time for temporal analysis.
How Does smFISH Work?
Single-molecule fluorescence in situ hybridization (smFISH) enables the visualization and quantification of RNA molecules at single-molecule resolution. This is achieved by hybridizing many short, fluorescently-labeled oligonucleotide probes to specific RNA targets. The technique is typically performed on fixed cells to ensure high specificity and spatial resolution.
How to detect mRNA with smFISH
- Design Probes: Create smFISH probes complementary to the target RNA sequence and label them with fluorophores for imaging.
- Prepare the Sample: Fix and permeabilize the sample to preserve cellular structures and allow probe access. While live-cell-compatible methods exist, they are experimental and not standard for smFISH.
- Hybridize Probes: Incubate the sample with smFISH probes under optimized conditions to ensure specific binding to the target RNA.
- Image Collection: Use the Vutara VXL microscope in large field-of-view (FOV) mode to capture high-resolution images of the labeled RNA molecules.
- Data Analysis: Analyze the collected data using Bruker's SRX software to localize, quantify, and visualize individual RNA molecules within cells.
Frequently Asked Questions
Understanding smFISH
Fluorescence in situ hybridization (FISH) is a technique used for the visualization of DNA, either specific genes or portions of genes, within a cell. This technique works via the use of probes that hybridize to the desired genomic region of interest and are labeled with fluorescent dye that is visible with SMLM. FISH is a powerful tool to investigate genomics questions, as it supports chromatin tracing applications. Like FISH, single molecule fluorescence in situ hybridization (smFISH) is a technique optimized for studying gene expression in single cells but is distinct in its ability to localize and quantify individual RNA molecules. With smFISH, RNA targets are imaged via the application of many short, labeled oligonucleotide probes that fluoresce when hybridized to the target sequence.
The advantage of our smFISH module is that it provides both visualization and quantification of RNA within the sample — unlike other techniques like RNA sequencing that only provide quantitative RNA abundance values.
Performing smFISH
The advantage of using Vutara’s smFISH module is that it provides both visualization and quantification of RNA within the sample — unlike other techniques like RNA sequencing that only provide quantitative RNA abundance values.
Advanced software is necessary to analyze smFISH data collected with SMLM. Bruker’s SRX software contains internal analysis tools, including those for segmentation and statistics, and is compatible with external open-source code. This software also features statistical analysis of distribution of smFISH probes in the cytosol and nucleus.
Bruker provides a variety of protocols for SMLM methods like smFISH. You can find them here.
An additional protocol for sample preparation and imaging with probes can be found here:
Trcek, T., Lionnet, T., Shroff, H. et al. mRNA quantification using single-molecule FISH in Drosophila embryos. Nat Protoc 12, 1326–1348 (2017). https://doi.org/10.1038/nprot.2017.030




