
DNA-PAINT Imaging
The Only Out-of-the-Box Solution for Multiplexed, Multicolor DNA-PAINT Imaging
Combined with Bruker's software-controlled, automated, fully-integrated Microfluidics Unit, the Vutara VXL super-resolution microscope is the only commercial system that offers out-of-the-box multiplexed DNA-PAINT capabilities. Performed on these tools, DNA-PAINT imaging allows for sub-10 nm localization precision in single-molecule localization microscopy with unlimited multiplexing potential.
Moreover, the Vutara microscope with microfluidics system provides the added benefits of:
- No-hassle setup with expert service and support to expedite time to data collection
- Results that are more reliable and repeatable than data collected with retrofitted systems
- Automation and multi-condition/multi-user programming for easily scalable experiment design
Find Out More About DNA-PAINT Solutions
Or contact us to learn more about performing DNA-PAINT experiments with the Vutara VXL microscope.
What is DNA-PAINT?
DNA-PAINT is a technique to enable single-molecule localization through the binding of short (<10 nucleotides) oligonucleotides labeled with a fluorophore to a complementary oligonucleotide bound to a target molecule, typically an antibody. It allows for extensive imaging modalities, from whole-cell extensive Z-stacks to high-resolution multi-target images.
Understanding the Advantages of DNA-PAINT Imaging
- Higher photon yield: The blinks are typically longer-lasting than in conventional dSTORM. This results in higher photon yields from the fluorophore and therefore allows for a much higher localization precision (<10 nm) when compared to such methods as dSTORM and PALM.
- Practically unbleachable imaging: The sample is bathed in an excess of fluorophore, allowing for extremely long-lasting imaging.
- Unlimited multiplexing potential: Since target specificity is set by nucleotide sequence, it is possible — with the right tools — to label multiple targets with different oligo sequences. With the Vutara’s integrated microfluidics unit, the imaging strand for a given target can be washed from the sample and different imaging strands labeling different biological targets can be added.
For further reading on Super-Resolution Microscopy with DNA-PAINT, see:
- Exploring DNA-PAINT
- Download the Knowledge Pack
- Single-Molecule Localization Microscopy—Beyond the Coverslip
- Single-Molecule Kinetics and Super-Resolution Microscopy by Fluorescence Imaging of Transient Binding on DNA Origami
- Multiplexed 3D Cellular Super-Resolution Imaging with DNA-PAINT and Exchange-PAINT
- Universal Super-Resolution Multiplexing by DNA Exchange
- Extended-Depth 3D Super-Resolution Imaging Using Probe-Refresh STORM
How Does DNA-PAINT Work?
DNA-PAINT works through the transient binding of a short “imaging oligonucleotide” containing a fluorophore to a complementary oligonucleotide — called the “docking strand” — on the target of interest, such as an antibody, nanobody, aptamer or suicide enzyme ligand.
Using Super-Resolution Microscopy with DNA-PAINT
Super-resolution imaging with DNA-PAINT involves several steps, including:
- The sample is labeled with the “docking strand” through conventional techniques and prepared for imaging.
- For imaging, the sample is bathed in imaging buffer (typically PBS but can include oxygen scavengers) and a low (typically 0.1-1 nM) concentration of imaging oligo complementary to the docking strand. The imaging oligo is typically 9-10 nucleotides in length and contains a fluorophore. We recommend Cy3B for DNA-PAINT due to its fluorogenicity and thus lower background.
- Once in the imaging buffer, the sample can be imaged. The transient binding of the imaging strand to the docking strand stops the diffusion of the fluorophore allowing it to be imaged on the camera.
Since the sample is bathed in a large excess of constantly exchanging imaging strand, the target is essentially unbleachable, making it possible to batch-process a large number of frames and extended Z-stacks.
Why Use DNA-PAINT for 3D Cellular Super-Resolution Imaging?
High-Precision Localization
DNA-PAINT allows sub-10 nm localization precision, making it one of the most precise microscope techniques available.
Here, a Vutara microscope with a water immersion 1.2 NA objective was utilized for a DNA-PAINT experiment. The image shows a whole BS-C-1 cell’s tubulin network labeled with tubulin antibodies conjugated to a DNA-PAINT secondary antibody. The inset shows a zoomed-in section of the tubulin network. The lumen of the microtubule is clearly visible.
Multicolor Unbleachable Imaging
Multiplexed multicolor super-resolution imaging is made possible with DNA-PAINT.
DNA-PAINT has the potential for large-scale multicolor Z-stacks due to the fact the sample is bathed in a practically limitless supply of fluorophore. This enables large-scale z-stack imaging composed of millions of localizations.
Here, a two-color DNA-PAINT experiment was performed on the Vutara single-molecule localization microscope. Tubulin is labeled in cyan and clathrin in magenta. Furthermore, due to the unbleachable nature of DNA-PAINT large Z-stacks are possible.
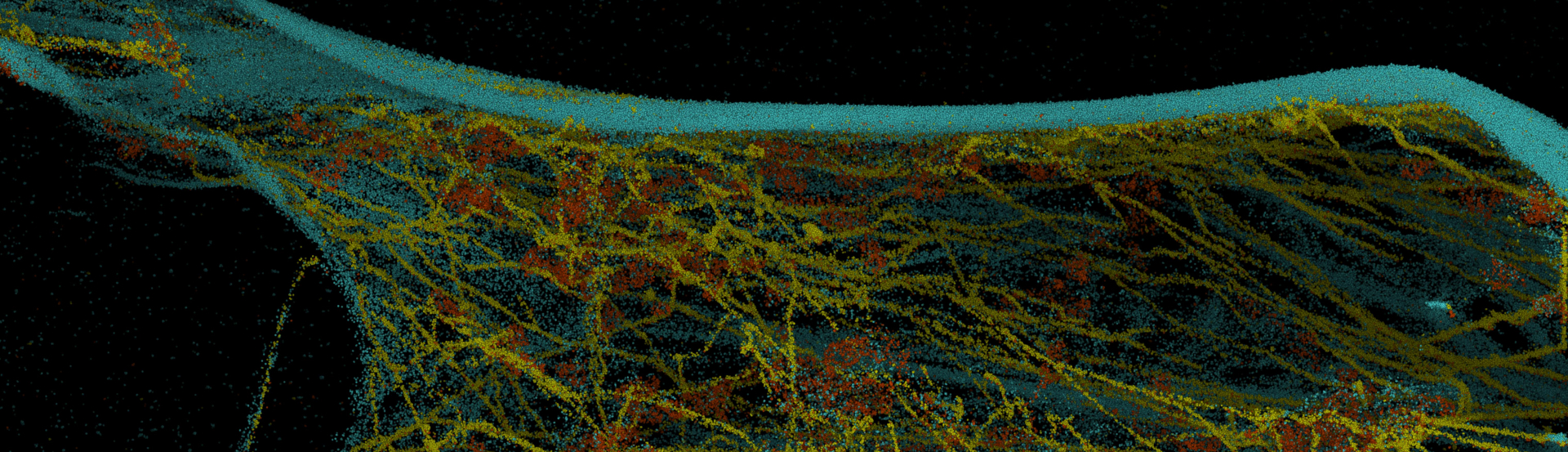
Unlimited Multiplexing Potential
DNA-PAINT has enormous potential for multiplexed imaging.
Here, a multi-target DNA-PAINT experiment was performed using the Vutara VXL and integrated fluidics unit. Using orthogonal docking strands on different probes, a potentially unlimited number of targets are possible.
Also pictured is a four-target DNA-PAINT experiment performed on the Vutara single-molecule localization microscope and integrated fluidics unit. F-actin-magenta, tom20-cyan, tubulin-yellow and clathrin-green.
Sample Images: DNA-PAINT for Improved Super-Resolution Imaging
BS-C-1 labeled with anti-tubulin, actin, anti-tom20 and anti-clathrin. Orthogonal 2º DNA-PAINT antibodies were purchased from Massive-Photonics.com.





